Current Projects
Current Projects in the Tran Laboratory
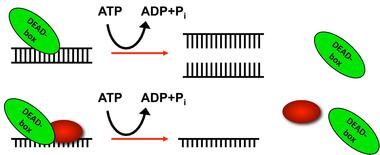
DEAD-box RNA helicases utilize ATP binding and hydrolysis to catalyze local changes to RNA structure and/or change RNA-protein composition.
Helicases are enzymes that bind and rearrange nucleic acid structures in vivo. These enzymes can act on DNA or RNA or mixtures of both. There are ~60 RNA helicases in mammalian cells, two-thirds of which are grouped into the DEAD-box RNA helicase family. Members of this class share the common, single letter amino acid motif D-E-A-D along with eleven additional conserved sequences and a common structural core. These enzymes undergo a cycle of ATP binding, RNA binding, ATP hydrolysis, and RNA release that is frequently coupled to structural changes in RNA and/or RNA-protein complexes/ DEAD-box RNA helicases are found in all domains of life with individual members functioning in every aspect of RNA biology from ribosome biogenesis to messenger (m) RNA decay, performing activities that are absolutely essential for cellular life. However, the precise targets for individual DEAD-box proteins in vivo are not well understood. Our lab is addressing this gap between biochemical characterization and biological function using the budding yeast model system S. cerevisiae. This research project involves a combination of enzymology and genome-wide approaches to understand the relationship between RNA structural changes and specific steps in RNA biology.
Cover of Molecular Cell showing that lncRNAs base-pair with DNA to control transcription. Cover by "resident artist" and co-author Siwen Wang.
Long non-coding RNAs (lncRNAs) are a heterogeneous class of RNAs that are greater than 200 nucleotides in length but lack an obvious open reading frame. Recent advances in RNA sequencing technologies lead to the identification of tens of thousands of these molecules in all eukaryotes from fungi to mouse to man. LncRNAs have been implicated in multiple, biochemically distinct processes in the cell including regulation of transcription of protein-coding genes, modulation of mRNA processing steps, and scaffolding multiple proteins together for assembly of macromolecular complexes. Our laboratory is investigating the mechanism of transcriptional regulation by lncRNAs and has proposed a model whereby these molecules control the timing of gene expression. We are currently testing this model using the S. cerevisiae model system and a combination of high throughput genetic screening strategies, high-resolution microscopy, and genome-wide approaches.
A surprising fact about RNA helicases is that many members of this enzyme class are upregulated in specific cancer types, suggesting a correlation between RNA helicase activity and uncontrolled cell growth. We have been working to understand how RNA helicases promote cancer and determine if this activity can be inhibited with small molecules. Our specific focus is on prostate cancer and lung cancer, due to clear links with specific DEAD-box RNA helicase gene amplifications and/or mutations. This research involves a combination of helicase enzymology, tissue culture, microscopy and genome-wide studies to define connections between RNA structural remodeling and uncontrolled cell growth.
