Research
Photosynthesis transforms the radiant energy of sunlight into usable chemical energy and thus sustains nearly all life on Earth. In plants and algae, this process takes place inside cytoplasmic organelles of endosymbiotic origin called chloroplasts. The thylakoid membranes within chloroplasts contain photosystems, pigment-protein complexes that capture sunlight and initiate photosynthetic electron transport reactions. Two distinct photosystems operate in series, connected by the cytochrome b₆f complex, to move electrons from water to NADP⁺, generating energy rich ATP and electron rich NADPH. These products of the light reactions fuel the Calvin-Benson cycle, which reduces CO₂ to carbohydrates. Our research investigates the genetic and molecular mechanisms that enable plants and algae to harness light efficiently and safely. Three major areas of investigation are described below.
Role of post-translational modifications in photosystem II function and biogenesis
Photosystem II (PSII) carries out the formidable water-splitting reaction of photosynthesis and thereby extracts electrons and protons necessary for the anabolic reactions of life. Incomplete water-splitting and or inadvertent electron transfer to molecular oxygen generate highly reactive oxygen free radicals that damage key protein subunits in PSII. PSII repair cycle is a highly orchestrated molecular process wherein the damaged PSII subunits are selectively degraded and replaced with newly synthesized copies while the undamaged subunits are simply recycled. How is plant PS II, a massive 1.4 MDa supercomplex embedded in the thylakoid membrane, disassembled for repair and reassembled after repair is an open question. Using biochemical and biophysical approaches we investigate the role of protein phosphorylation and oxidative protein modification in PS II disassembly and repair. Our investigations are expected to provide insight into the molecular mechanisms that ensure the orderly disassembly and repair of plant PSII.
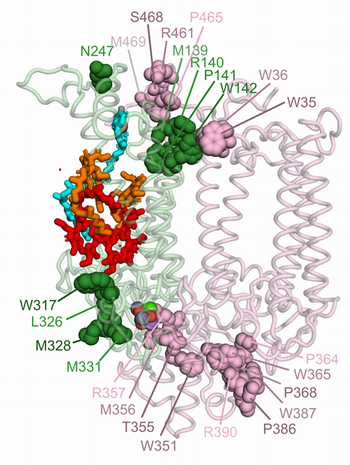
Oxidative modifications of D1 and CP43 subunits are likely critical for plant PSII disassembly. Oxidatively modified residues of D1 are highlighted as dark green spheres, and those of CP43 as dark pink spheres. We hypothesize that these modifications at the D1-CP43 interface facilitate the dissociation of the PSII monomeric core into a repair-compatible RC47 complex and a CP43-precomplex.
Function and signaling mechanisms of Chloroplast Sensor Kinase
Chloroplast Sensor Kinase (CSK) is a bacterial-type sensor kinase first identified in Arabidopsis as a putative regulatory link between photosynthetic redox reactions and chloroplast gene expression. Our lab recently demonstrated that CSK uses an iron-sulfur cluster to sense the redox state of plastoquinone (PQ), a key photosynthetic electron carrier. In cyanobacteria and non-green algae, CSK appears to function within a classical two-component system to regulate heat-shock response genes upon sensing the PQ redox signal. In plants and green algae, however, chloroplast heat-shock genes have relocated to the nucleus, and CSK has lost both its response regulator partner and its conserved histidine autophosphorylation site. CSK nevertheless appears to regulate chloroplast genes. Our research seeks to understand how plant and algal CSK have acquired new functions and signaling mechanisms.
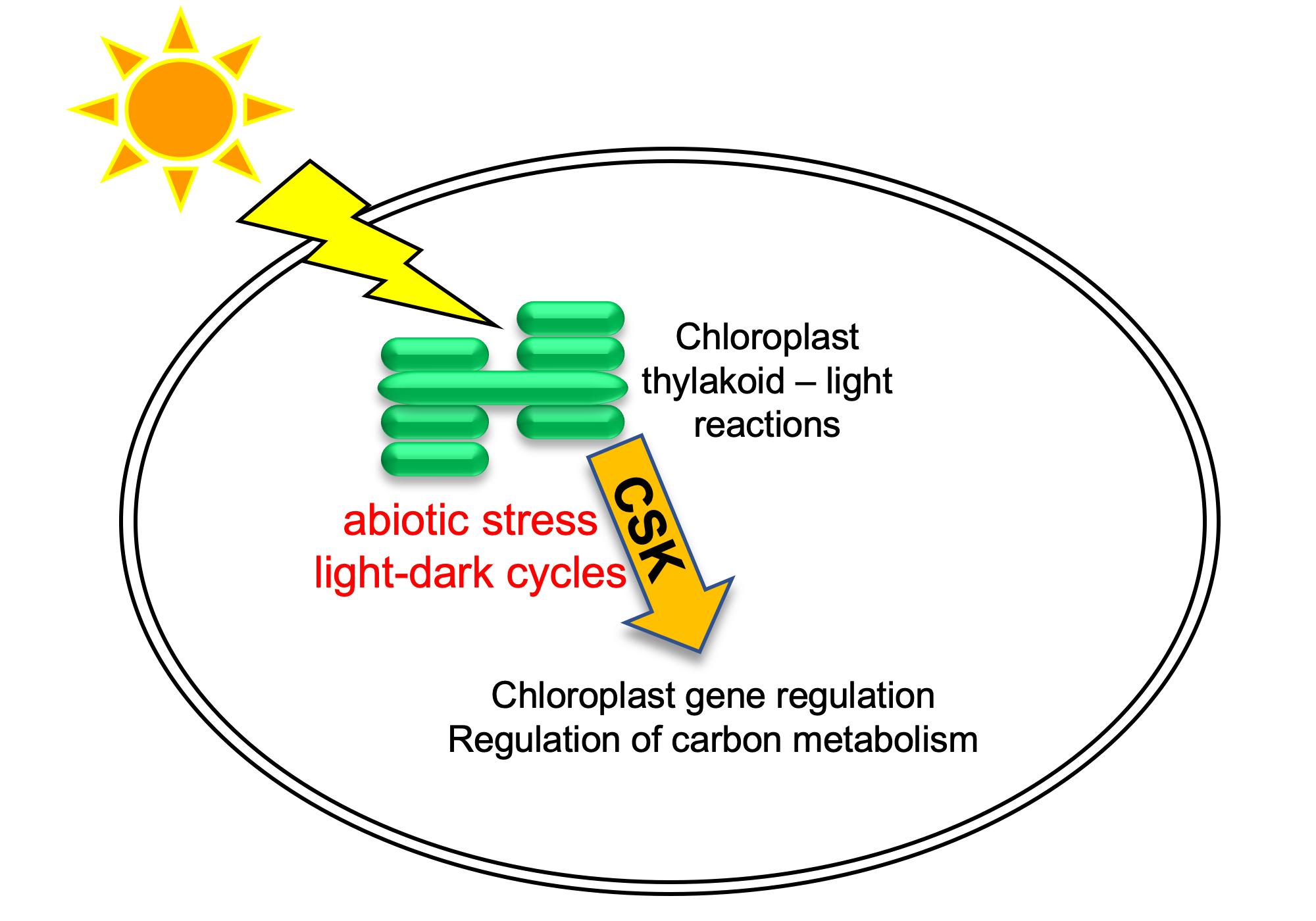
A model for CSK function in chloroplasts. Abiotic and diurnal signals influence photosynthetic light reactions, which are subsequently sensed by CSK. CSK then modulates chloroplast gene expression and carbon metabolism to optimize photosynthesis under prevailing environmental conditions.
Diatom photosynthesis under changing light and nutrient conditions
Diatoms are a group of mostly aquatic, photosynthetic algae with plastids of red algal ancestry. They fix as much carbon as all rainforests combined. We use the model diatom Phaeodactylum tricornutum to decipher the genetic basis of their high photosynthetic efficiency under changing light and nutrient conditions.
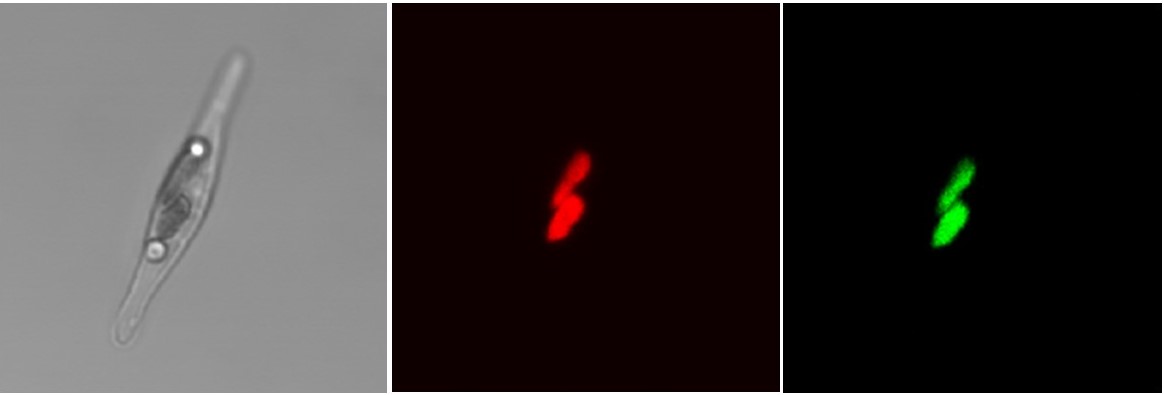
Plastid-localization of Phaeodactylum Chloroplast Response Regulator 1 (CRR1). Far-left panel is a bright field image of a Phaeodactylum cell with a dividing chloroplast; the middle panel, chlorophyll autofluorescence; and far-right panel, green fluorescence from CRR1-GFP fusion protein. (In collaboration with Stefan Zauner and Uwe Maier, Philipps University of Marburg)
