RESEARCH
Biological functions are ultimately determined by proteins. The levels and activities of proteins are controlled by both protein synthesis and protein degradation. In eukaryotic cells, the majority of protein degradation is performed by the ubiquitin-proteasome system (UPS). Proteins selected for degradation are tagged with the small protein ubiquitin, a process referred to as ubiquitination, and proteins tagged by a chain of four or more ubiquitin are subsequently recognized by the 26S proteasome for degradation (Fig. 1). Ubiquitination is crucial for a broad spectrum of essential physiological processes in both plants and animals. Accumulated evidence has shown that misregulation in protein ubiquitination can lead to various human diseases, or defects in plant growth and disease defense.
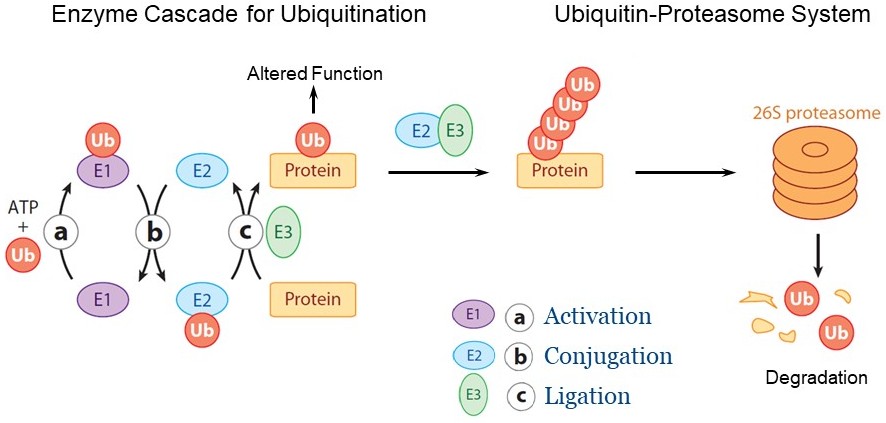
Figure 1 The ubiquitin-proteasome system and the enzyme cascade for ubiquitination. Ubiquitination is achieved by a cascade of three sequentially-operating enzymes comprising ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligating enzyme (E3). Ubiquitin is first activated by E1 and is then transferred and conjugated to E2 (E2~Ub). The E3 binds both the substrate protein and the E2~Ub, enabling the transfer of ubiquitin to the substrate. Substrate proteins with a chain of four or more ubiquitin are degraded by the 26S proteasome, while proteins with a mono-ubiquitin usually display altered functions.
The goal of our research is to understand the role and mechanism of protein ubiquitination. Currently, our major research effort is to investigate the largest family of ubiquitin ligases, cullin–RING ligases (CRLs). These enzymes determine the substrate specificity of the ubiquitination reaction, and they are typified by the SKP1•CUL1•F-box protein (SCF) E3 ligases. Similar to other CRLs, SCFs are composed of a Cul1 scaffold protein and one of the many interchangeable SKP1•F-box protein substrate receptor modules that recognizes specific substrates (Fig. 2). Therefore, recruiting the right type of F-box protein at the right time is essential for proper SCF function, and how the cellular repertoire of SCFs is dynamically controlled to meet the needs of a cell to target a changing pool of substrates for ubiquitination has been a critical question.
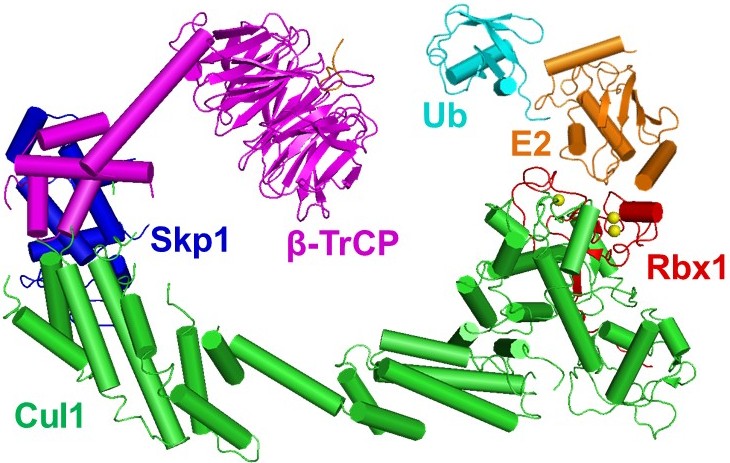
Figure 2 Structure of a SCF complex. CUL1 of SCF recruits the F-box protein substrate receptors through the adapter SKP1. SCF recruits E2 via the RING protein RBX1.
To help answer this fundamental question, we have proposed and provided evidence for an “adaptive exchange” model: the limited pool of CUL1 constantly and rapidly scans the entire population of F-box•SKP1, ensuring timely recruitment of substrate bound F-box•SKP1 and the subsequent degradation of the substrates. This highly dynamic process is enabled by Cullin-associated Nedd8-dissociated protein 1 (CAND1), an F-box protein exchange factor, and Nedd8, a ubiquitin-like protein that modifies cullins (Fig. 3).
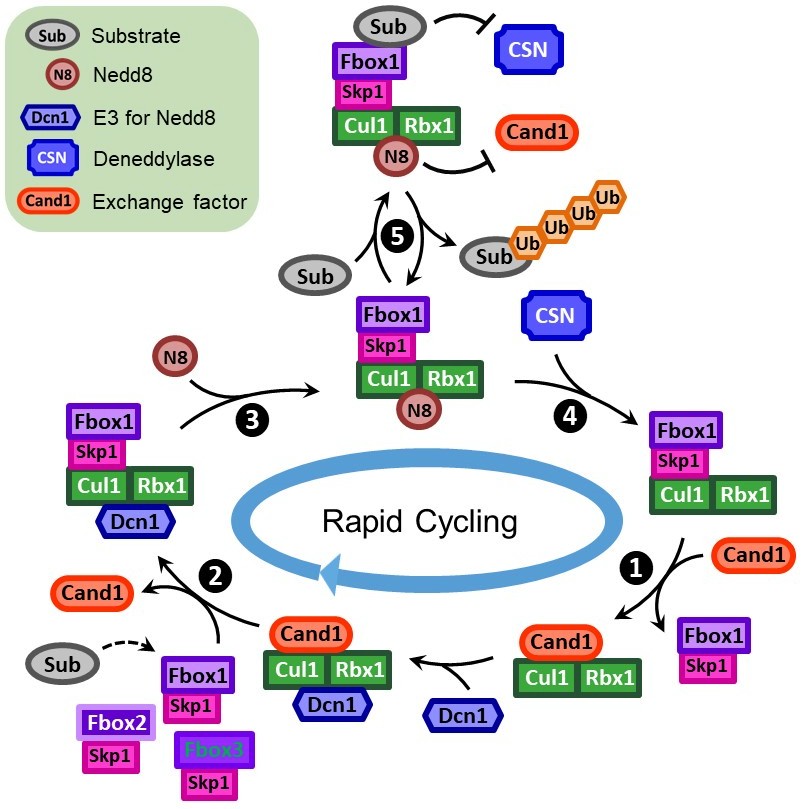
Figure 3 Simplified model for the rapid cycling of CUL1. CAND1 dramatically increases the dissociation rate of an existing SCF and binds tightly to the CUL1 “recycled” from the SCF (Step 1). Subsequently, a new F-box•SKP1 can then destabilize the CUL1•CAND1 complex, remove CAND1, and form a new SCF (Step 2). This exchange reaction is controlled by Nedd8 modification: immediately after the removal of CAND1 by F-box•SKP1, CUL1 is neddylated and can no longer be exchanged by CAND1 (Step 3); only when the NEDD8 is cleaved by the COP9 signalosome (CSN), the CUL1 is subject to CAND1-mediated exchange again (Step 4). Furthermore, substrate binding inhibits the NEDD8 deconjugation by CSN, allowing the formation of an SCF that is stable enough to ubiquitinate the substrate (Step 5).
With a wide array of approaches such as biochemistry, biophysics, cell biology, quantitative proteomics, and molecular genetics, we are studying the mechanism and regulation of the CRL network. We are now working to expand our knowledge of CRLs from CUL1-based SCF complexes to other cullins in the CRL family. We also aim to study the relative roles of CAND1 and CAND2 in human cells.
