CRACKING THE
MICROBIOME CODE
CRACKING THE
MICROBIOME CODE
CRACKING THE
MICROBIOME CODE
Thousands of years ago, the first farmers learned how to harness the natural world around them and cultivate it to provide food, clothing, medicine and energy. They observed wild plants and animals and learned the rules governing how they grow, reproduce and interact. Then they used that knowledge to raise heartier, more nutritious crops, breed animals for desirable traits, and harvest medicines and fuel from natural sources.
Today, the same process is restarting on a much smaller — and yet infinitely broader — scale. Research on the microbiome, the invisible ecosystem of bacteria, viruses and other microorganisms all around and even inside of us, has intensified in the past decade as scientists use new genetic methods to survey these complex populations. Now, as the field shifts from cracking the microbiome code to manipulating and engineering it for human and environmental benefits, Purdue scientists are breaking new ground in cultivation at the microbial scale.
Purdue researchers are rewiring microbes to create the biofuels and drugs of tomorrow and determining which foods can create a healthier human or animal gut. They’re creating platforms and biosensors to test and monitor new treatments that engineer the microbiome in humans, animals, plants and soil. Their laboratories utilize gene editing, nanotechnology and other cutting-edge approaches to figure out how the microbiome quietly mediates many natural processes we take for granted — and how it can be redirected to make both humans and the world we live in healthier.
 And as they study the richly interwoven communities of the microbiome, they’re creating one of their own on the Purdue campus, bridging departments and areas of expertise and sharing ideas with the potential to reshape multiple fields. Microbiome research is typically found in medical centers and biology departments, not in a college of agriculture. This unique setting gives Purdue’s microbial experts a holistic perspective, as they reveal how the global microbiome serves as the hidden interface between more visible forms of life on earth.
And as they study the richly interwoven communities of the microbiome, they’re creating one of their own on the Purdue campus, bridging departments and areas of expertise and sharing ideas with the potential to reshape multiple fields. Microbiome research is typically found in medical centers and biology departments, not in a college of agriculture. This unique setting gives Purdue’s microbial experts a holistic perspective, as they reveal how the global microbiome serves as the hidden interface between more visible forms of life on earth.
Feeding your inner ecosystem
Public acceptance of the microbiome can be tracked by the rise of the probiotic. It wasn’t long ago that most people lumped all bacteria and their microbe peers into the blanket category of “germs,” using antibiotics and antibacterial products to eradicate them without prejudice. Today, as we begin to understand that many microbial species work with us, not against us, probiotic goods have proliferated in the form of pills, yogurt, kimchi and other grocery aisle staples, typically billed as promoting a healthier digestive system.
One big problem, says Stephen Lindemann, assistant professor of food science, is that science still doesn’t quite know what a “healthy” human gut microbiome looks like, never mind how a probiotic helps realize it. Researchers have focused on cataloging and characterizing the microbiomes of various environments, essentially taking a sample of genes and using the information to take attendance of the species present in the population. Scientists can then compare these populations in different groups — say, people with celiac disease and people without — but have learned little about which particular populations cause or prevent illness, and how.
“Without that kind of mechanistic understanding we can’t really tune the microbiome to do what we want it to do,” Lindemann says. “We’re stuck in a world where we can only say this output is correlated with health or unhealth, or with respect to a specific condition. There’s nothing actionable there.”
So in Lindemann’s lab, they’re digging into how the human gut microbiome handles a common but important set of nutrients: dietary fibers. Our own gastrointestinal system can’t fully digest many of these complex carbohydrates, so we rely on our microbial tenants to convert them into usable components. Lindemann’s group tests how different fiber sources affect the composition and activity of the microbiome. They found that even physical grain features such as bran particle size can produce radically different effects.
“The problem — and opportunity — is that we have found that, with respect to the structure of dietary fiber, everything matters,” he says.
 By examining these responses, he hopes to find new ways to use diet to influence the microbiome, nudging these ecosystems toward healthier outcomes with the right mix of cereal grains for maximum benefit. Farther in the future, he envisions “designer carbohydrates,” with plants bred and cultivated for their microbiome-enhancing properties and custom fiber diets for individuals based on their specific microbial needs and responses.
By examining these responses, he hopes to find new ways to use diet to influence the microbiome, nudging these ecosystems toward healthier outcomes with the right mix of cereal grains for maximum benefit. Farther in the future, he envisions “designer carbohydrates,” with plants bred and cultivated for their microbiome-enhancing properties and custom fiber diets for individuals based on their specific microbial needs and responses.
“It’s really important we think about microbiomes as ecosystems, because they are full of interactions,” Lindemann says. “Our goal is to try to come up with the principles that guide the ecology of the gut, so that we can then use those principles to engineer or manage it.”
 Harnessing the animal microbiome
Harnessing the animal microbiome
That vision for human health can easily be extended to animals, where the diets and digestive microbiome may differ, but many of the goals are the same. Tim Johnson, assistant professor of animal sciences, has looked at the proliferation of antibiotic-resistant microbes in livestock that are fed antibiotics to promote growth and prevent illness. Animal producers are looking for ways to raise their livestock without antibiotics, turning to diet and the microbiome as a potential solution.
“In the past, the approach was to suppress bacterial growth in the gut. But that’s going to suppress both pathogenic and beneficial bacteria — microbes in the gut that stimulate the immune system and prevent disease,” Johnson says. “So now the idea is, instead of relying on antibiotics, let’s actually try to promote the gut microbiome and make use of all the beneficial bacteria that are there to promote health.”
In collaboration with animal nutritionists and livestock producers, Johnson’s lab researches new feed additives that can produce these desired effects in swine and poultry. Starting with the basic corn or soybean-based diet of these animals, they’re adding in different complex carbohydrates and closely observing how the microbiome responds: how its population of microbial species shifts, and how its generation of different beneficial nutrients, such as fatty acids, changes.  After those relationships are established, the right mix of additives could be a viable, and more sustainable, alternative to feed antibiotics.
After those relationships are established, the right mix of additives could be a viable, and more sustainable, alternative to feed antibiotics.
“The end goal is to reduce our need to treat animals with antibiotics and hopefully to reduce levels of antibiotic resistance as well,” Johnson says.
Beyond the gut
Many animal microbiomes do things that their human counterparts can’t. Ruminants, including cattle, giraffes and sheep, have strategies to break down grass, leaves and other plants that we find indigestible. To do so, fungal members of their gut microbiome produce powerful enzymes for extracting nutrition from plant matter.
These digestive enzymes, if successfully harvested and controlled, could be an incredible boost for biotechnology, says Kevin Solomon, assistant professor of agricultural and biological engineering. Plants absorb atmospheric carbon dioxide and fix it in their leaves, stems and roots; modern biotech approaches seek to convert that rich biomass into sugars that serve as the building blocks for biofuels, medicines and other chemical products. But today’s processes are limited in the kinds of plant material they can break down, Solomon says.
 “Plant biomaterial is a complex polymer with different sugars that are interlocked together, and each of those chemical bonds you can think of as a lock,” Solomon says. “Current technologies produce a lot of keys, but only for a handful of those locks. The family of fungi we study has an enormous diversity of enzymes, so we can actually produce 100 different types of keys, break apart more types of bonds, and release more free sugar from a material. The question is, ‘How do we mass-produce those enzymes, and how do we get the cells to produce them on demand?’”
“Plant biomaterial is a complex polymer with different sugars that are interlocked together, and each of those chemical bonds you can think of as a lock,” Solomon says. “Current technologies produce a lot of keys, but only for a handful of those locks. The family of fungi we study has an enormous diversity of enzymes, so we can actually produce 100 different types of keys, break apart more types of bonds, and release more free sugar from a material. The question is, ‘How do we mass-produce those enzymes, and how do we get the cells to produce them on demand?’”
To do so, Solomon’s group isolates these fungi from the dung of various ruminants — common livestock as well as more exotic species such as zebras, giraffes and rhinos — then uses the gene-editing tool CRISPR to “rewire” their enzymatic activity. The goal is to create enzymes that can break down otherwise useless agricultural waste into the ingredients for several valuable products. Think of it like a brewery, Solomon says, where giant vats contain chemical reactions producing bioethanol or antimalarial drugs, instead of beer.
Creatures great and small
On the other end of the animal spectrum from the rhinoceros is the tiny aphid, which Assistant Professor of Entomology Laramy Enders calls “the most adorable and destructive insect.” Within these tiny pests, there’s a microbiome with important effects on both the aphids themselves and the plants they devour. The microbial communities inside an aphid could determine, for example, whether it carries the harmful barley yellow dwarf virus or can provide protection against the toxic defense of the milkweed plant.

Decoding the aphid microbiome adds new insight into the fascinating co-evolution, over millions of years, of plants, their insect predators and the microbes that live on and inside both groups. But it could also provide new targets for farmers looking to protect their crops without the use of chemical pesticides.
“We’re looking for ways to engineer or use the microbiome against an aphid,” Enders says. “Can we take advantage of or manipulate some of the interactions between the viruses and other microbes found inside aphids? Or can we introduce other microbes to compete with and disrupt some of those interactions?”
Soil provides yet another microbiome that could play a key role in this plant–insect battle. With Professor of Entomology Ian Kaplan, Enders is studying whether the microbial community in soil can be engineered to help plants defend themselves from insect pests. Although the project is in its early stages, she envisions a potential system where farmers submit soil samples that are assessed by scientists, cultivated to possess a protective microbiome and sent back for “seeding” their home fields.

But even on its own, the relatively simple network of the insect microbiome offers a useful prototype for the engineering approaches eventually intended for humans and their far more complex communities.
“I think they’re quite powerful in their simplicity and ability to be experimentally understood and manipulated. We can use insects to really get at some fundamental processes happening in microbial communities,” Enders says. “That can then potentially extend to other systems and help our general understanding across lots of different types of microbiomes.”
A thriving research culture
When all is well in the ecosystem of the microbiome, each species has its own complementary role and strengths, and nobody works in isolation. The same could be said of the Purdue microbiome research community, which has bloomed with the introduction of several new faculty in the last two and a half years.
“We have a group of highly engaged, highly interactive young faculty who are really excited to tear up these problems in really similar ways in very different systems,” Lindemann says. “Our community is kind of like the microbiome in some ways: We are highly interactive, and that provides research output greater than the sum of its parts.”
The strength of that team, as well as the enormity of their chosen research area, was expressed by a symposium the group organized last spring, which linked the behavior of tiny microbes to global challenges in human and animal health, economic growth and environmental sustainability.
 That’s not hyperbole. Just as the dawn of agriculture dramatically altered human culture and behavior, harnessing the microbiome could also reshape society. Many proposed innovations would recycle waste, protect endangered habitats, reduce the overuse of antibiotics and toxic pesticides, and produce other environmental benefits. Solomon even sees the microbial biotechnology to grow new fuel sources and medicines in vats as a politically stabilizing force, easing tension around limited or fragile natural resources.
That’s not hyperbole. Just as the dawn of agriculture dramatically altered human culture and behavior, harnessing the microbiome could also reshape society. Many proposed innovations would recycle waste, protect endangered habitats, reduce the overuse of antibiotics and toxic pesticides, and produce other environmental benefits. Solomon even sees the microbial biotechnology to grow new fuel sources and medicines in vats as a politically stabilizing force, easing tension around limited or fragile natural resources.
Being part of the College of Agriculture provides the broader context and expertise necessary to address these challenges, the faculty agree. Instead of focusing exclusively on human-centered, medical applications of the microbiome, they’re assessing its role in the environment, food systems, and other places where nature and humanity are deeply entangled.
Faculty are also forging new microbiome collaborations with researchers in the colleges of Engineering, Health and Human Sciences, Science and Veterinary Medicine. These projects combine disciplines and skill sets to increase our understanding of the role of microbiomes in shaping diverse systems. One example is using mathematical models to predict the results of experiments in which gut microbes consume different carbohydrate structures. Another is predicting whether microbiomes in wetland sediments will give rise to high methane emissions.
“You want to be in a place where you have a thriving agricultural program that you can plug into this new aspect, which is considering how microbiomes are important for agricultural sustainability,” Enders says. “Our researchers are moving away from the idea that you have these silos of just looking at the plant microbiome, or the insect microbiome, or humans, and we’re beginning to integrate across those different core areas of microbiome science. It just feels more holistic, and it’s the type of community that you want if you really want to push everything forward.”
MICRO SYSTEMS,
NANO TOOLS

To observe and influence organisms too small for the naked eye to see, you need tools that are even smaller. Enter Mohit Verma, assistant professor of agricultural and biological engineering, whose background in nanotechnology enables him to create devices that study the interaction between microbes and host organisms. Here are some of the tools he’s using in the lab to examine the microbiome of the infant gut.
NANOPORES: Many microbes grow well in a lab dish; host cells are less sturdy. Left to their own devices, the former would quickly overwhelm the latter.
So Verma separates them with nanopores, with a diameter near one millionth of a centimeter, which let nutrients through but not the microbes themselves.
GUT-ON-A-CHIP: Body organs typically consist of multiple cell types with different functions; organs-on-a-chip attempt to simulate this diversity in a three-dimensional culture. Verma uses a “gut-on-a-chip” platform that replicates the human intestine, allowing researchers to introduce various microbes under different conditions and observe the response of this artificial digestive tract.
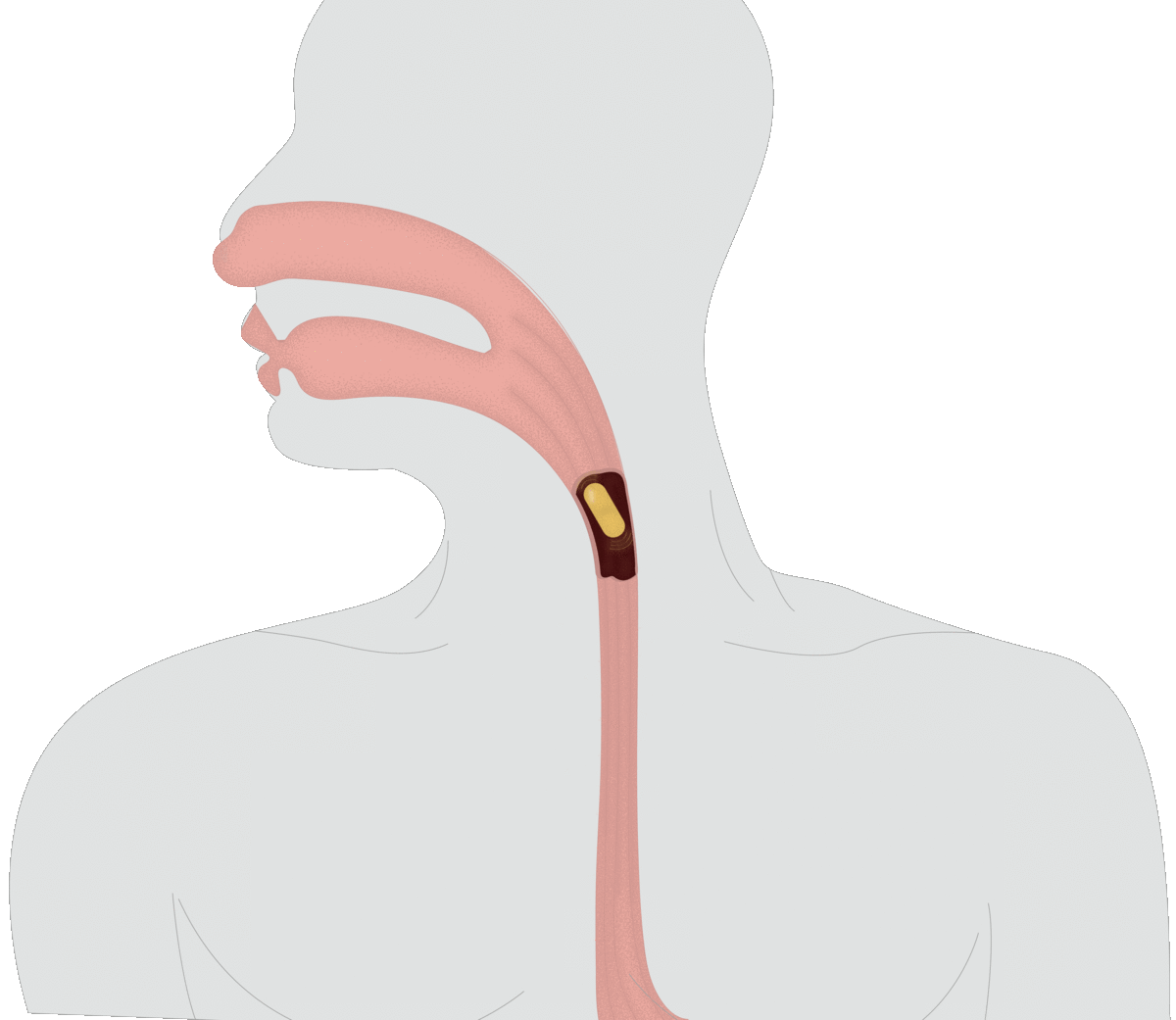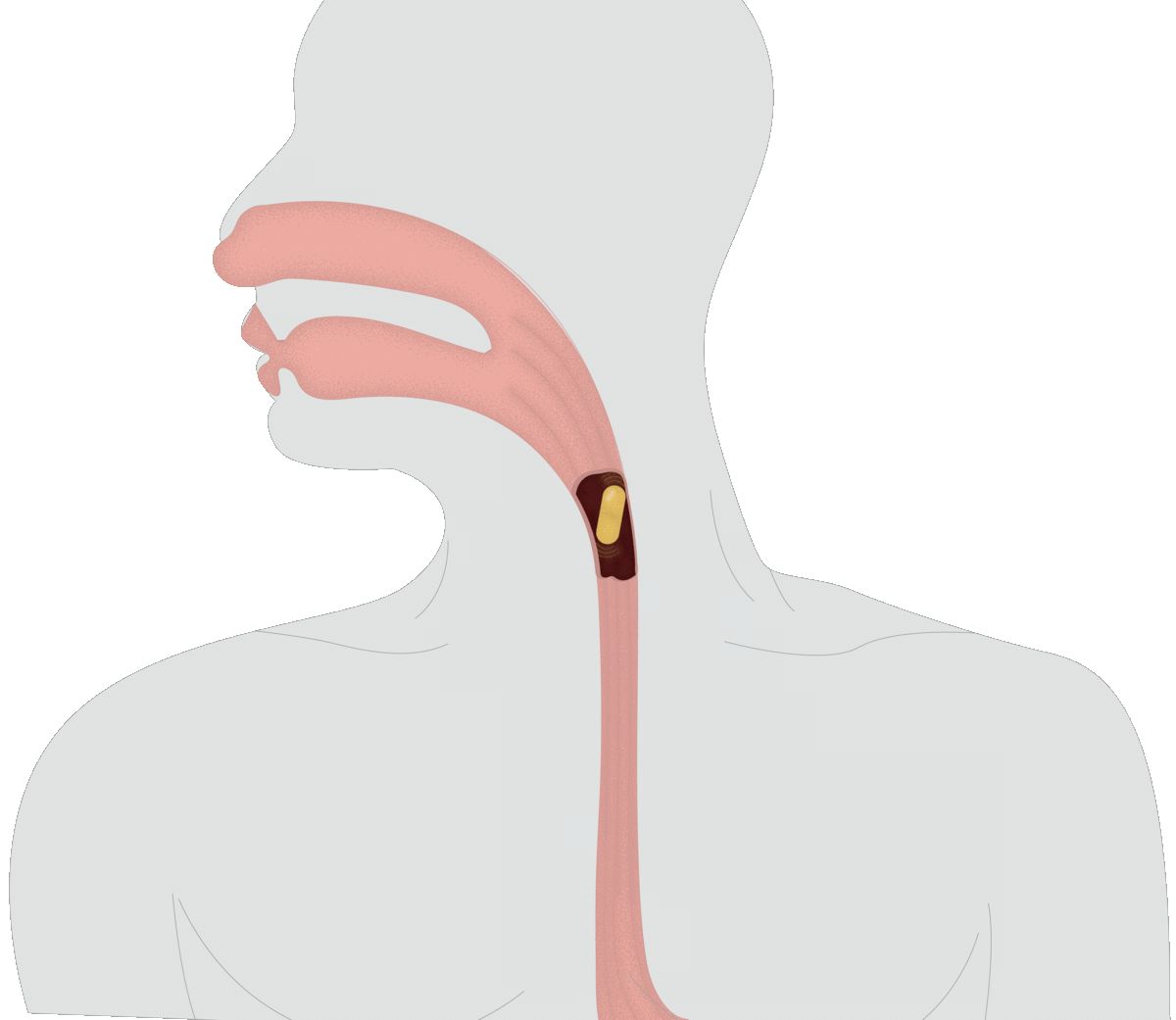
SMART CAPSULES: Currently, the best noninvasive way to assess an individual’s microbiome is dirty work: taking a fecal sample. With Assistant Professor of Materials Engineering Rahim Rahimi, Verma is inventing a better way to collect data on the microbiome, in the form of a swallowable pill. As the capsule moves through a human or animal digestive system, it will transmit data on microbial abundance, inflammation and other measures to help scientists monitor the microbiome in real time.
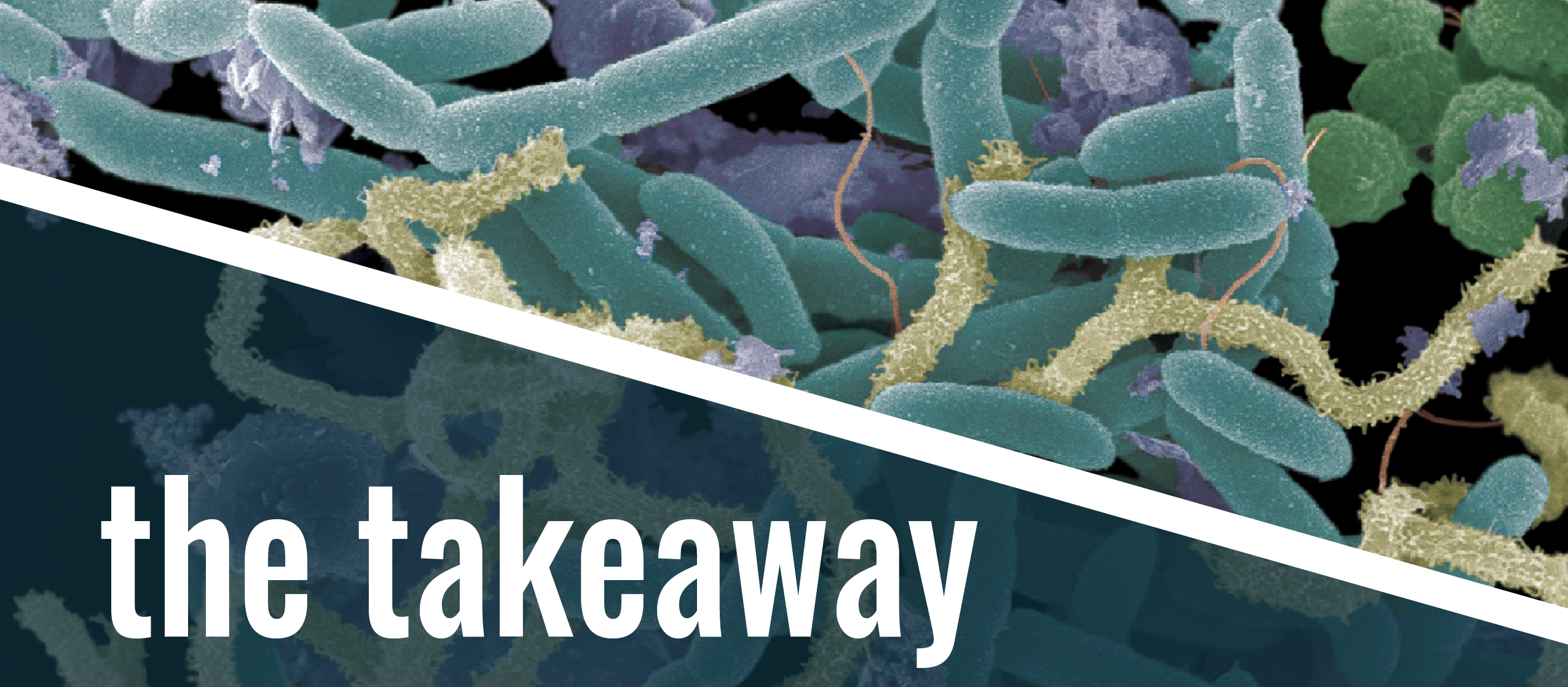
Banner photo: More than the sum of its parts: A three-membered microbial consortium from soil (Agromyces spp. [yellow cells], Micrococcus spp. [green cells] and Paenibacillus spp. [teal cells]) requires a symbiotic relationship to grow when the environmental pollutant dibenzofuran [purple crystalline forms] is provided as the only source of carbon and energy. Photo by Tim Johnson; colorized by Lauren Coghlan.
Purdue Agriculture, 615 Mitch Daniels Blvd, West Lafayette, IN 47907-2053 USA, (765) 494-8392
© 2024 The Trustees of Purdue University | An Equal Access/Equal Opportunity University | USDA non-discrimination statement | Integrity Statement | Copyright Complaints | Maintained by Agricultural Communications
Trouble with this page? Disability-related accessibility issue? Please contact us at ag-web-team@purdue.edu so we can help.