Research Overview
The Lindemann lab researches mechanisms by which diet influences the gut microbiome and the effects of those microbial shifts on host health, including inflammation and gut colonization by pathogens. The goal is to discover dietary solutions to health problems such as Crohn’s disease, diabetes, inflammatory bowel disease, colorectal cancer, ulcerative colitis and more. Specifically, the Lindemann lab focuses on using molecular ecology techniques to understand how diet influences the composition and stability of the gut microbiome; how gut microbiome metabolism of dietary components influences production and absorption of bioactive microbial metabolites; how metabolic interactions between microbes alter polysaccharide fermentation and nitrogen metabolism in the colon; and how these interactions between beneficial microbes exclude pathogenic organisms and modulate inflammation in the colon.
Projects
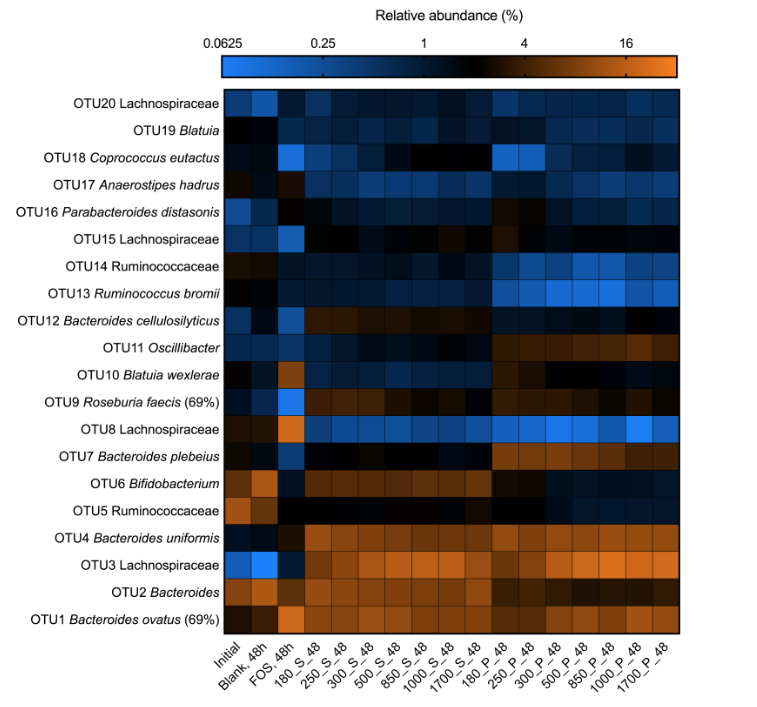 Large grain particles often go undigested through the intestinal tract. Around the world, many grains are not consumed as large grains, but instead as finely processed powders, allowing for enhanced digestibility. The human gut microbiome is largely responsible for degrading grain particles – including diverse natural polysaccharides that are unavailable to human enzymes. Very little is known regarding the influence of particle structure and size on gut ecology and function. This project aims to determine whether the differences in grain bran size select in vitro for different microbial communities from the same fecal inoculum, and whether phylogenetically linked organisms have species level substrate-dependent differences. Ultimately, we are aiming to identify genomic variances that allow similar organisms to differently utilize the soluble (supernatant) and insoluble (particle) substrates. Towards this goal, we are using metagenomic analysis to distinguish relevant putative proteins involved in substrate-based selection. A combination of contig assembly, genome binning, and relative abundance calculations allow for the observation of informative genomes whose comparisons shed light on their respective resource preferences.
Large grain particles often go undigested through the intestinal tract. Around the world, many grains are not consumed as large grains, but instead as finely processed powders, allowing for enhanced digestibility. The human gut microbiome is largely responsible for degrading grain particles – including diverse natural polysaccharides that are unavailable to human enzymes. Very little is known regarding the influence of particle structure and size on gut ecology and function. This project aims to determine whether the differences in grain bran size select in vitro for different microbial communities from the same fecal inoculum, and whether phylogenetically linked organisms have species level substrate-dependent differences. Ultimately, we are aiming to identify genomic variances that allow similar organisms to differently utilize the soluble (supernatant) and insoluble (particle) substrates. Towards this goal, we are using metagenomic analysis to distinguish relevant putative proteins involved in substrate-based selection. A combination of contig assembly, genome binning, and relative abundance calculations allow for the observation of informative genomes whose comparisons shed light on their respective resource preferences.
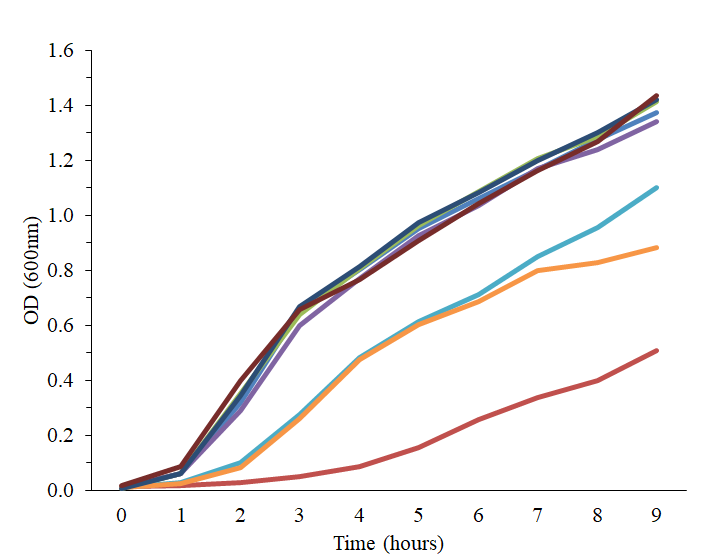 Genome encoded properties are responsible for microbial competition for carbohydrates. Glycoside hydrolases, while responsible for metabolism of a given polysaccharide do not allow for long chain-length polymers to be digested without competition. Therefore, it is likely that differences in transport strategies and hydrolase location likely influence the outcome of microbial competition for a resource. In vitro cultivation and evolution of strains capable of metabolizing dietary fibers with varying sizes combined with whole genome sequencing will determine responsible genetic features. Future experiments will combine computational modeling, chemical biology with probes, and high throughput screening of host impact to determine ecological relationships and carbohydrate structures.
Genome encoded properties are responsible for microbial competition for carbohydrates. Glycoside hydrolases, while responsible for metabolism of a given polysaccharide do not allow for long chain-length polymers to be digested without competition. Therefore, it is likely that differences in transport strategies and hydrolase location likely influence the outcome of microbial competition for a resource. In vitro cultivation and evolution of strains capable of metabolizing dietary fibers with varying sizes combined with whole genome sequencing will determine responsible genetic features. Future experiments will combine computational modeling, chemical biology with probes, and high throughput screening of host impact to determine ecological relationships and carbohydrate structures.
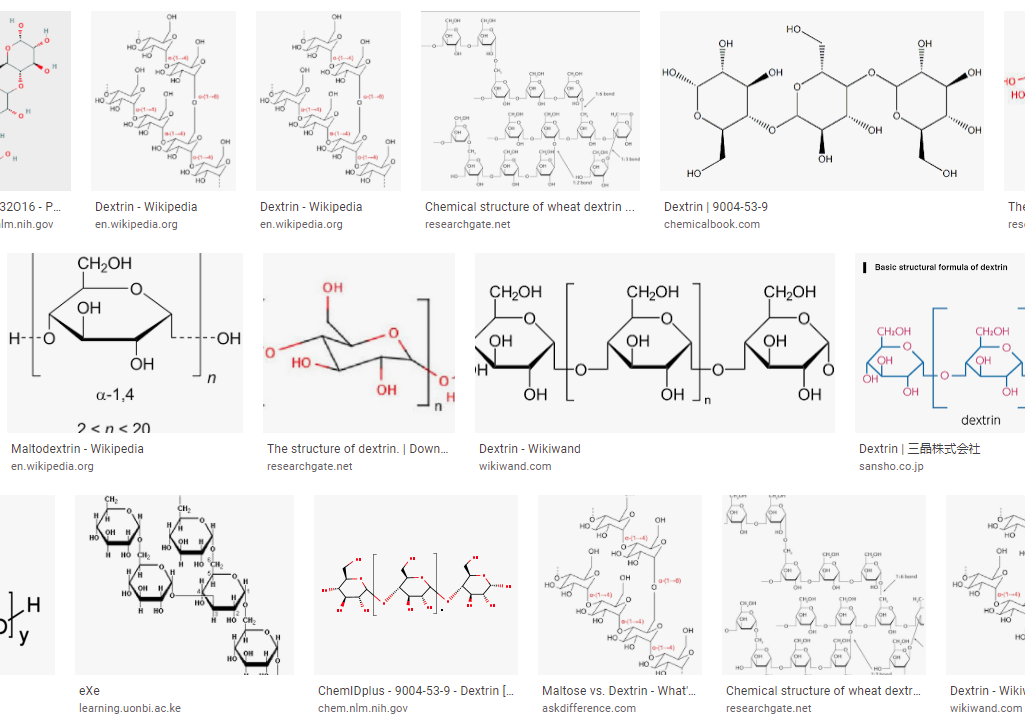 Dietary fibers are typically plant carbohydrates that cannot be digested by the human system. Dextrins are a class of dietary fibers that are made of D-glucose units linked by α-(1→4) or α-(1→6) glycosidic bonds and can be created during human digestion, malting and mashing, or during the baking of bread. While abundant research is available on the simple substrate or mixed polysaccharide consumption by microbial communities, little is known regarding the effect of defined complex substrates on these same communities. This project aims to determine the effect of increasingly complex substrates with the same fundamental building blocks on microbial ecology. Specifically, intestinal microbial communities fed with dextrins of varying molecular weights, degrees of polymerization, and linkage types, can be observed for community shifts and selection after both extended incubation and passage.
Dietary fibers are typically plant carbohydrates that cannot be digested by the human system. Dextrins are a class of dietary fibers that are made of D-glucose units linked by α-(1→4) or α-(1→6) glycosidic bonds and can be created during human digestion, malting and mashing, or during the baking of bread. While abundant research is available on the simple substrate or mixed polysaccharide consumption by microbial communities, little is known regarding the effect of defined complex substrates on these same communities. This project aims to determine the effect of increasingly complex substrates with the same fundamental building blocks on microbial ecology. Specifically, intestinal microbial communities fed with dextrins of varying molecular weights, degrees of polymerization, and linkage types, can be observed for community shifts and selection after both extended incubation and passage.
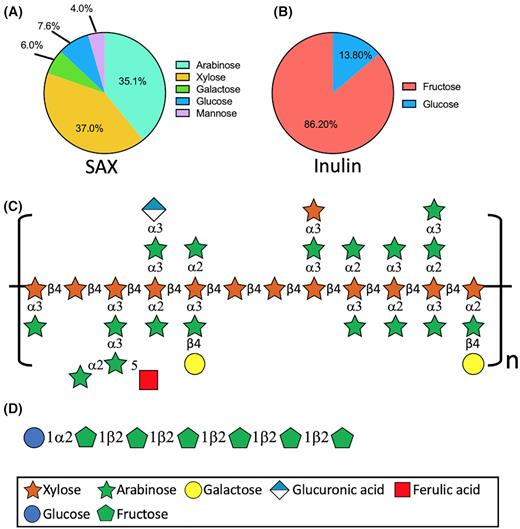 Extending on work completed with the dietary fiber dextrins, arabinoxylans also provide a vehicle for understanding how fine differences in fiber chemical structure leads to structural and functional shifts in gut microbiota composition in vitro. This project aims to resolve the microbial species level differences and rationally employ different AX structures to shape microbial community structure and function. Specifically, correlating the physical, chemical, and structural characteristics of arabinoxylans with the metabolic fate of these fibers and the microbial community will reveal members responsible for and enriched by specific diets.
Extending on work completed with the dietary fiber dextrins, arabinoxylans also provide a vehicle for understanding how fine differences in fiber chemical structure leads to structural and functional shifts in gut microbiota composition in vitro. This project aims to resolve the microbial species level differences and rationally employ different AX structures to shape microbial community structure and function. Specifically, correlating the physical, chemical, and structural characteristics of arabinoxylans with the metabolic fate of these fibers and the microbial community will reveal members responsible for and enriched by specific diets.
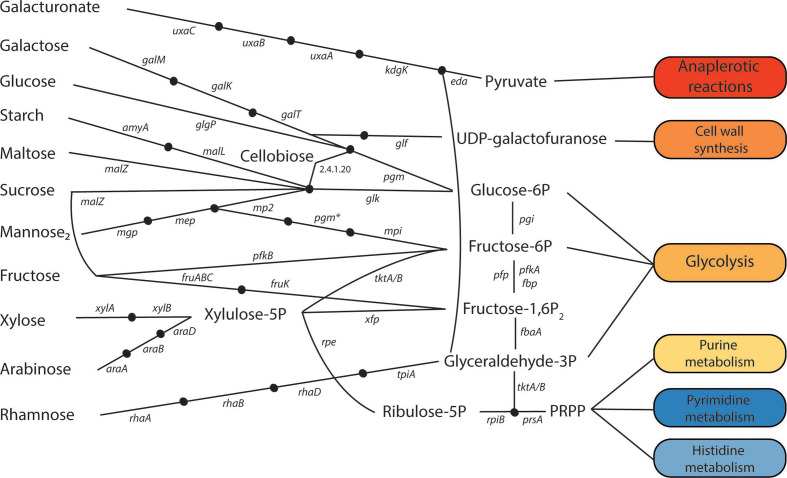 Hoping to understand the ecological principles governing consumption of oligosaccharides by communities. Specifically, seeking to identify how differential gene content and regulation among members influence the outcomes of competition for oligosaccharides in communities, and, in turn, host physiology. Projects under this purview entail construction of dynamic compositional and metabolic models to simulate and predict the outcomes of interactions among community members of oligosaccharide-fermenting consortia. Utilizing metagenomic and genome reconstruction, multi-omic functional analysis, and metabolic modeling models will be tested to relate microbial gene content / regulation and competitive advantage on diverse carbohydrate substrates.
Hoping to understand the ecological principles governing consumption of oligosaccharides by communities. Specifically, seeking to identify how differential gene content and regulation among members influence the outcomes of competition for oligosaccharides in communities, and, in turn, host physiology. Projects under this purview entail construction of dynamic compositional and metabolic models to simulate and predict the outcomes of interactions among community members of oligosaccharide-fermenting consortia. Utilizing metagenomic and genome reconstruction, multi-omic functional analysis, and metabolic modeling models will be tested to relate microbial gene content / regulation and competitive advantage on diverse carbohydrate substrates.
 We do fun stuff. Join us!
We do fun stuff. Join us!
The mission of the Lindemann Lab is to train well rounded conscientious scientific leaders with the passion, ability, and persistence to transform the world and to generate the knowledge and tools required to predict and control microbiomes for human health, economic benefit, and environmental sustainability. Our chief aim is to build exceptional leaders, from that, exceptional research follows. If you are interested in joining the Lindemann Lab, please send your CV and a cover letter to Dr. Stephen Lindemann.

