New assay accelerates E. coli testing process
A team led by Purdue University’s Bruce Applegate has developed a new time-saving assay to detect an especially severe strain of E. coli in ground beef.
The toxin produced by E. coli accounts for 175,000 illnesses annually in the United States. The U.S. Department of Agriculture’s Food Safety and Inspection Service has maintained zero tolerance for O157:H7, an E. coli strain that can be life-threatening, since 1994. Zero tolerance means that if even one cell of E. coli is detected in a standard 325-gram (11.4-ounce) sample of ground beef, the entire batch is flagged as unfit for consumption.
“The issue is, you need to have a detection technology where you can find one cell in your sample,” said Applegate, a professor of food science. But one E. coli cell is almost impossible to find in a 325-gram sample. “There’s not a technology able to do that unless you do what we call an enrichment.”
The enrichment process entails preparing a special environment that allows E. coli O157:H7 to outcompete all other microorganisms in a food sample before testing.
“Instead of one cell in this 325-gram sample, I will have 10 million to 100 million of these cells in there. I can guarantee I will find it then,” Applegate said.
The assay both enriches the sample and can detect E. coli during the 15 hours or more needed to ship samples from a production facility to a testing laboratory of the Food Safety and Inspection Service. Applegate and five co-authors published the details of their assay in the journal Foods.
The new assay saves time by combining enrichment and detection during shipment.
“Having an accurate reading in a very short amount of time means everything to not just the food industry, but safety as a whole,” said Chuyan Chen, co-lead author of the Foods paper. Chen, who received her master’s degree in food safety and microbiology at Purdue in 2018, now works in the consumer packaged foods industry. Chen noted that the work can be adapted for application to other food pathogens as well.
“We know that O157:H7 isn’t the only microorganism that can get humans sick. There are many others,” she said.
Claudia Coronel-Aguilera, another co-lead author and a former postdoctoral researcher in Applegate’s lab, now works as a research scientist in the dairy industry.
“For us, time to detection is the main problem. The faster you have results, the faster you release the product,” Coronel-Aguilera said. “You’re using enrichment during transportation, so when the product arrives, you can already have a positive or negative result.”
The new assay has promising implications for nations in Africa, South America and elsewhere that have fewer resources than the U.S. to invest in food safety.
“The testing methods that we have in the U.S. are expensive,” Coronel-Aguilera said. But the method described in the Foods paper is a fast, easy and much less expensive version of the highly effective lab methods. “That will be a game changer,” she said.
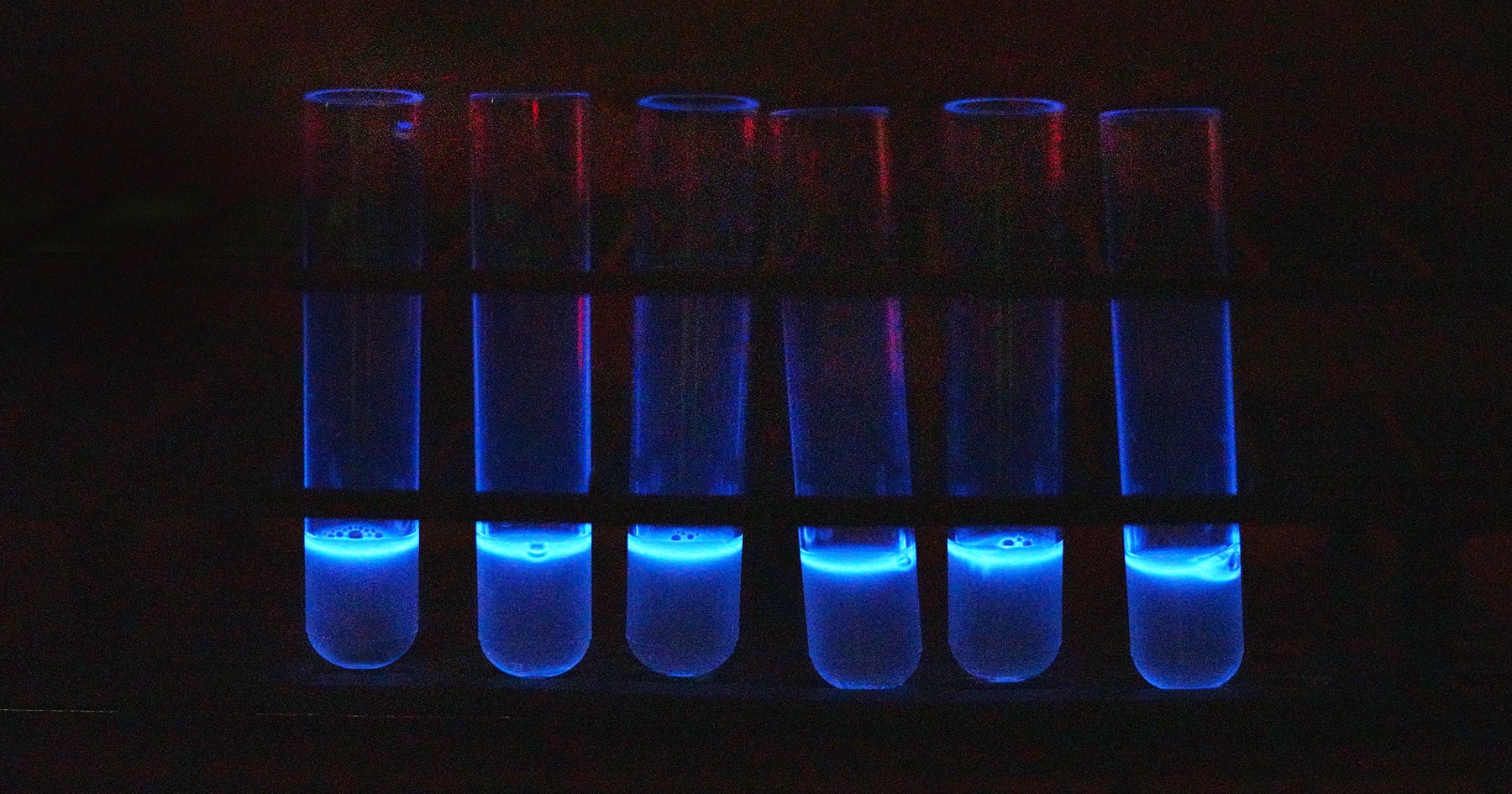 New, faster test for pathogenic E. coli in beef is based on using a modified phage, a virus that infects bacteria. After infecting the pathogenic E. coli, the phage produces an enzyme that causes the bacteria to glow.
New, faster test for pathogenic E. coli in beef is based on using a modified phage, a virus that infects bacteria. After infecting the pathogenic E. coli, the phage produces an enzyme that causes the bacteria to glow. The system is based on phages — viruses that infect specific bacteria.
“We genetically modified this phage in such a way that it would infect the bacteria and cause it to produce light,” Applegate said.
After infecting E. coli O157:H7, the phage integrates its genome into the bacterial cells’ chromosome. Once integrated into the E. coli O157:H7 chromosome, the phage produces an enzyme that makes light and causes the infected cells to glow. A glowing culture indicates the presence of the pathogen.
“Then you’re making more bacteria glow over time. It’s like shingles in a way,” Applegate explained. “You get chickenpox. You get blisters from the dying cells, but also that virus gets into your cells and then you get shingles later on.”
The talk Chen gave about the work won the Best Oral Presentation Award at the 20th International Symposium on Bioluminescence and Chemiluminescence in 2018 in Nantes, France. The phage, she noted, is specifically evolved to infect only one type of bacterium.
“Our phage is not specific to any E. coli, only to E. coli O157:H7,” she said.
Additional co-authors of the Foods paper include Arun Bhunia, a Purdue professor of food science, and Andrew Gehring and George Paoli of the USDA’s Agricultural Research Service in Wyndmoor, Penn. Gehring and Paoli’s collaborations with Applegate are part of a longstanding cooperative agreement between the USDA’s Agricultural Research Service and Purdue’s Center for Purdue Food Safety Engineering.