Breaking down the biochemical pathways of protein degradation
Proteins are more than just important nutritional units to track in your diet. They are encoded into our DNA and are the building blocks of all living things, making up everything from the lettuce leaves in your salad bowl to the fingers holding your fork. Some proteins even help activate chemical processes, like the processes you’ll need to break down that salad later today.
One significant research topic in the world of protein is its degradation. Protein degradation is important for gene expression regulation, cell signaling, repair after exposure to adverse environmental conditions and protection from malformed proteins that can be toxic.
Within the established machinery for protein degradation, Xing Liu and her lab in the Department of Biochemistry study the pathway called ubiquitination. Ubiquitination is named for the protein ubiquitin, which acts as a tag for protein degradation. In this pathway, ubiquitin is passed through a series of enzymes—those proteins that activate chemical processes—until it gets to one called an E3 ligase that allows it to attach to a protein the cell is targeting for degradation.
A chain of ubiquitin hanging off of another protein signals the need for that protein to be delivered to the proteasome that will break it down. This tag placed by the E3 ligases allows ubiquitination to do something special: provide specificity to the process of protein degradation. Instead of breaking down everything in one area of the cell, it only breaks down precisely which proteins are marked.
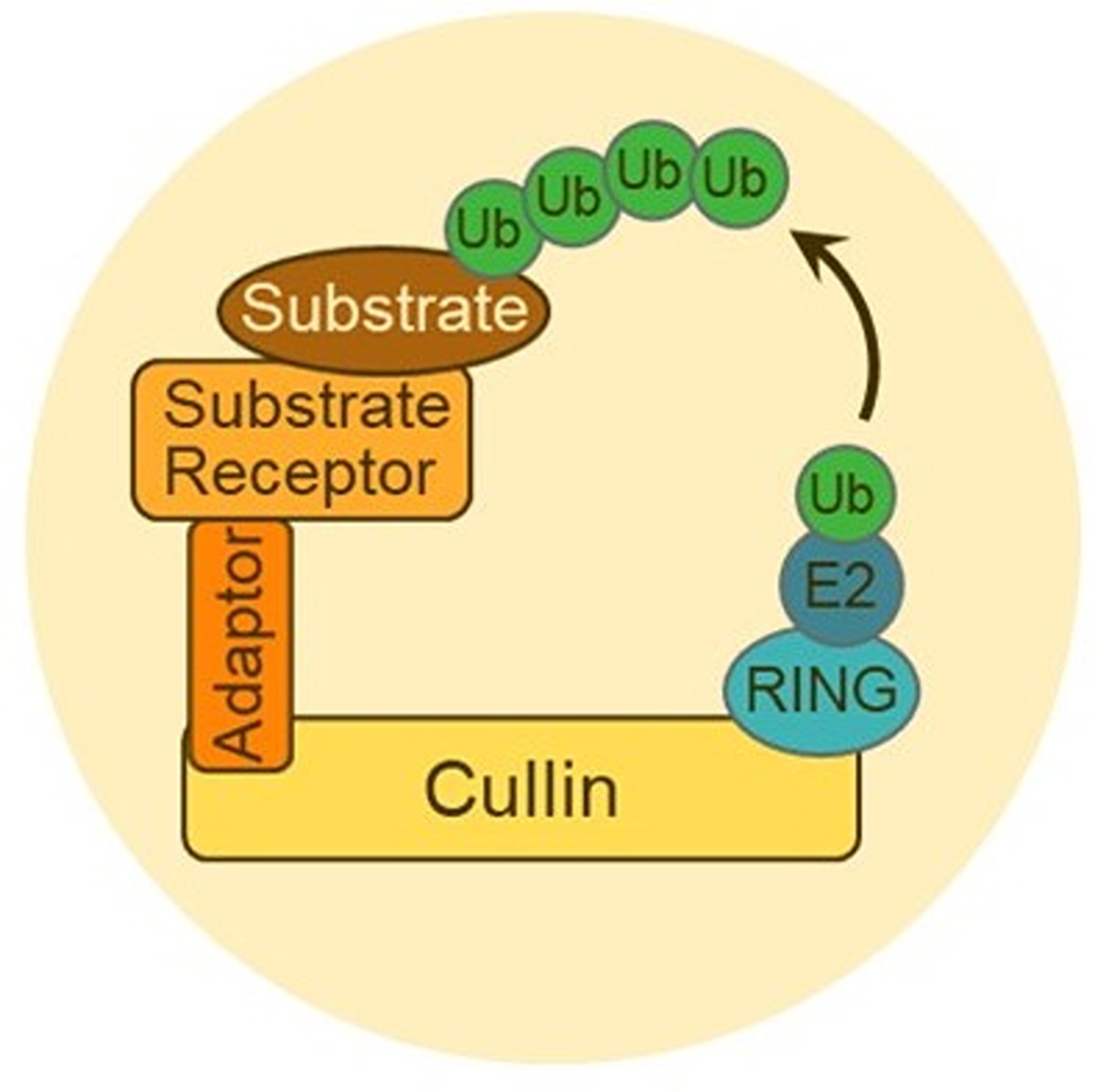
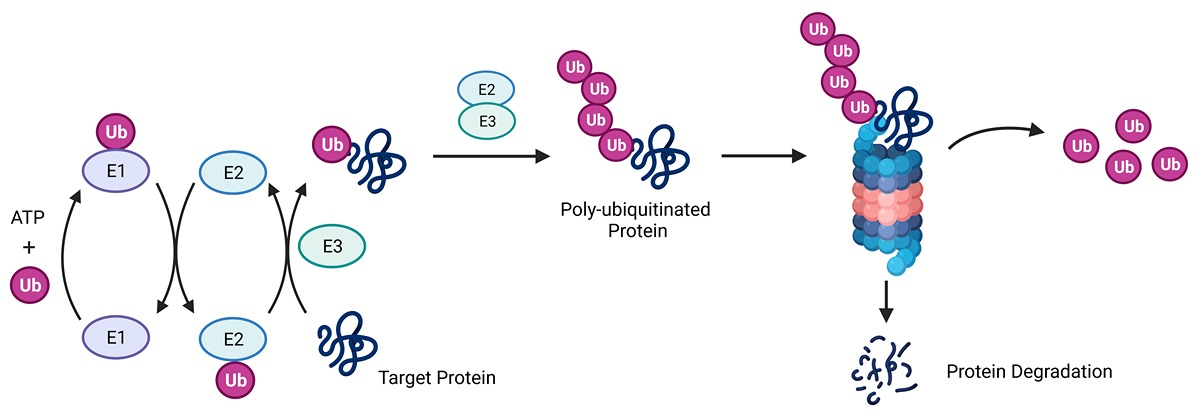 In the process of ubiquitination, ubiquitin (Ub) is passed through a series of enzymes, and an E3 ligase attaches it to a target protein. Once a target protein has several ubiquitin attached in a chain, it goes to a proteasome that degrades it, and the ubiquitin is recycled. Diagram provided by Liu lab.
In the process of ubiquitination, ubiquitin (Ub) is passed through a series of enzymes, and an E3 ligase attaches it to a target protein. Once a target protein has several ubiquitin attached in a chain, it goes to a proteasome that degrades it, and the ubiquitin is recycled. Diagram provided by Liu lab. Liu and her postdoctoral trainee Kankan Wang have been studying the E3 ligases to unravel their role in protein specificity. There are around 600 E3 ligases in humans, and Liu and Wang study the largest family, called the Cullin-RING Ligases (CRLs). CRLs have major impacts on pharmaceutical development and play a role in fighting various diseases, including cancer, by breaking down toxic proteins.
“CRLs make such a large family, not just because they have diverse protein components, but also because they have something special in their structure,” Liu explained. “It’s a protein complex that contains multiple proteins. The Cullin is the bigger protein, and it has a banana shape. On one end of the banana-shaped Cullin, there is a small RING protein. The other end of the Cullin attaches to interchangeable complexes of substrate receptors.”
In humans, there are six different types of Cullins (CULs), and Liu lab studies CUL1, the first to be discovered, and CUL2, which is important for tumor suppression. Different CULs interact with different sets of interchangeable substrate receptors (SR), each of which is able to bind to different, specific proteins to be tagged. This is how the CRLs are selective towards the proteins they mark for degradation.
In previous studies, it was discovered that a protein called CAND1 binds to all the different CULs. Liu found that CAND1 wraps around CUL1 to displace the already-associated SR and detaches when a new SR comes to interact with CUL1. CAND1’s binding and detachment allows the SRs to more quickly and dynamically switch between each other, making it an exchange factor for the CUL1 protein. Through accelerating the dynamic assembly of new CRLs, CAND1 effectively activates CUL1, making it more efficient towards its specific target.
CUL2 is a very different story. Liu and Wang were able to discern that CAND1 did not increase the speed of protein degradation when it interacted with CUL2. It did the opposite—the targeted proteins took even longer to degrade than normal.
Liu and Wang’s data in their paper, published in Nature Structural & Molecular Biology, dives deeper into the interaction between CUL2 and CAND1.Liu shared, “We think this inhibitory effect of CAND1 on CUL2 is important because it can help distinguish the substrates with different affinities for the CUL2 system. If we think of a normal, natural substrate, it may already have some affinity with your CRL complex, but it's pretty weak, so it is still doing its job instead of getting ubiquitinated and degraded. But, when you have some regulation like phosphorylation or the attachment of molecular glue, that enhances that affinity. CAND1 then becomes like an affinity gate.”
While protein degradation is an important part of our natural biochemical systems, degrading all proteins constantly would leave us without the necessary building blocks and signals that we need to survive. So, CAND1 provides a way to make sure that the target proteins attaching to the CUL2 system are ones that actually should be degraded. If they have too low affinity for their specific SR, then they can probably stick around the cell and still do good work.
Liu has been part of CRL research since graduate school, where she studied auxin, a plant hormone that functions as a molecular glue for CRLs.
I'm just curious about what's going on in these organisms. I was so interested in plants because, unlike animals, they cannot move away from anything bad to them. So how do they deal with tough situations? They’ve got to have some mechanism to keep themselves alive and fit, and I wanted to know what happens at the molecular level that guides them through challenges.”
- Xing Liu, Assistant Professor of Biochemistry