Same shape, same function, different performance—a tale of two proteins
Every organism on Earth is a well-oiled machine in its own environment. It makes sense then that every piece of every process within all beings would be optimized to withstand generations of evolutionary pressure. If it survives, it must then be the fittest. In biochemistry, however, scientists often discover puzzling cases of inefficient actions in organisms: expenditures of extra effort and energy for something seemingly redundant.
Xing Liu and her lab in Purdue’s Department of Biochemistry share this story in their latest publication in Nature Communications. They studied selective protein degradation, or how cells get rid of and recycle obsolete, malfunctioning or excess proteins in the cell that act as active catalysts,regulators and general materials.
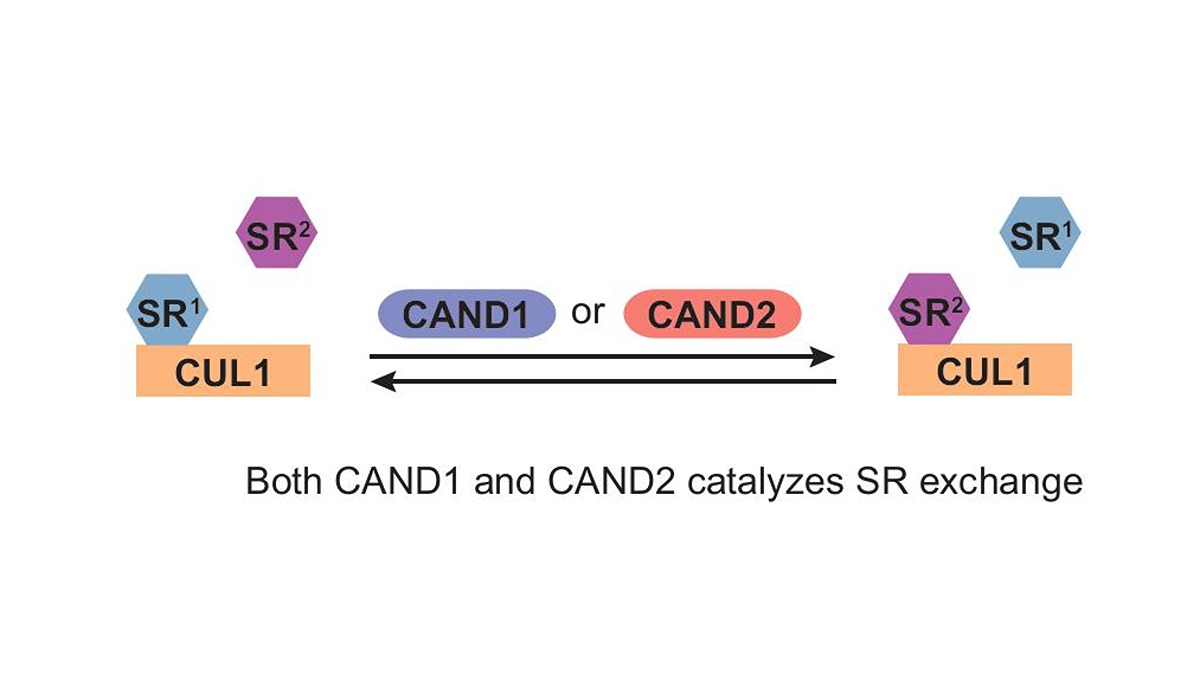
Even the process of protein degradation is run by proteins. The family of cullin proteins, or CULs such as the CUL1 used in this study, partner with Substrate Receptor (SR) proteins to identify other proteins that need to be broken down. Together, this CUL-SR complex adds ubiquitin protein tags on identified proteins, similar to how stores add clearance stickers over a discontinued item they need to move off the shelves.
CUL-SR complexes need help switching their focus between which proteins in the cell need to be removed. “The CUL1-SR complexes themselves are very stable, which becomes a challenge because CUL1 is supposed to change between the 69 different SRs it partners with in human cells to target different proteins for degradation. This should be a dynamic process.” Liu said.
Liu’s previous work discovered that the protein CAND1 helped the CUL1 protein take off one SR and switch it for another, thus allowing it to recognize and tag different proteins. This exchange factor is so important it exists in nearly all eukaryotes, including plants and animals. Without it, the CUL1-SRs would struggle to keep up with the overstock of proteins in the cell, including damaged and malformed ones that could cause harm. CUL1-SRs have roles in tumor suppression for this reason, and CAND1 makes them more efficient and effective.
CAND1, however, has an elusive sibling: CAND2. This protein consists of a highly similar amino acid sequence. Liu and her collaborator, Chittaranjan Das—a professor of chemistry in the College of Science—built the molecular structures of CAND2 to find that it looks nearly the same as CAND1. It also can attach to the same CUL1-SR complexes. It even performs the same function: kicking one SR off and getting another  one attached. However, CAND2 is predominantly found in mammals.
one attached. However, CAND2 is predominantly found in mammals.
“If you look at the Human Protein Atlas, CAND1 is almost everywhere—in all kinds of cell types. It's actually an essential gene. If you knock it out of mice, it's lethal.” Liu said. “CAND2 is highly enriched in just a few different tissues, including heart muscle cells and skeletal muscle cells.”
Knocking CAND2 out of mice is not lethal. In humans, however, it is linked with obesity and atrial fibrillation, an irregular heartbeat. When Liu’s lab removed CAND2 from human cells, they found the degradation of hundreds of proteins had been altered.
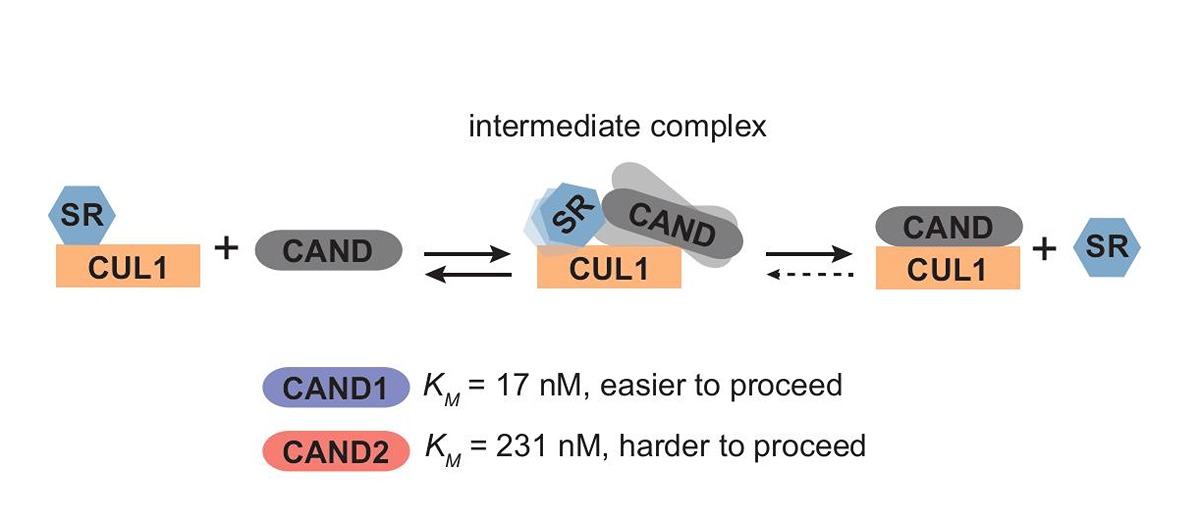
Even stranger, Liu found that CAND2 is not nearly as efficient as CAND1. Using a stopped-flow machine, her lab was able to measure the rate of each step of the SR exchange by CAND1 and CAND2 to compare them. The major difference they found was in the measurement of KM, which describes the affinity of the intermediate complex formed when a CAND1 or CAND2 protein attaches to the CUL1-SR. That affinity is significant in CAND1 or CAND2 actually completing the task of knocking off one SR and allowing another to attach.
CAND2 has a higher KM value, and thus its intermediate is harder to form, so it cannot take off or add an SR as successfully as CAND1. You would need a greater concentration of CAND2 proteins at work to accomplish the same amount of SR exchanges as CAND1.
So, why would a protein whose function is already carried out more efficiently by another be necessary in a few cell types in a few groups of organisms?
Liu and her lab are still working to answer this and other questions. Currently, most of their work on CUL-SRs, CAND1 and CAND2 proteins has been done in established human cell lines, which are not specialized to the particular cell types where CAND2 is prominent. Liu would like to continue the investigation in heart and skeletal muscle cells, to see how CAND2 performs differently from CAND1 where it is naturally found. She’s also been collaborating with Yun Zhou in botany and plant pathology on confocal microscopy, in which they can stain and view where CAND1 and CAND2 are compartmentalized within the cell, a common regulatory mechanism controlling protein activities.
Future studies on the structure of CAND2 seek also to figure out how it joins with CUL-SRs, changes confirmation in the intermediate complex, and how these processes contribute to its activity. These molecular features, combined with knowledge of where the protein is expressed and how much of it is expressed within the cell, may inform why proper activity of CAND2 is required for healthy cells.
Liu received the Maximizing Investigators’ Research Award (MIRA) from NIH to support her research program, and she also won the American Heart Association Career Development Award for her work to affect the future of heart health. “Curiosity about the fundamental knowledge of nature and life is the drive for my research. The biological questions of how these proteins work are my focus, and knowing that what I do has implications for disease makes those questions even more interesting.”