Purdue awarded $2.3 million NSF grant to improve cotton fiber engineering
The U.S. grows high-yielding types of cotton that have fibers that are thicker and shorter than their luxury pima cotton relative from Egypt. Improving the shape and mechanical properties of cotton cells would make this already $25 billion industry more valuable for U.S. growers.
Purdue University cell biologist Dan Szymanski, the principle investigator for the $2.3 million National Science Foundation grant, believes he can accomplish this by manipulating the cell walls of the higher-yielding G.hirsutum cottons. The traits Szymanski’s team will engineer are controlled by complex cascades of molecular and genetic processes, but simplified by cotton fibers being single cells that emerge from the developing seed coat. His team will analyze gene regulatory circuits, protein complex dynamics and cell wall properties as a function of fiber development to discover the control modules of fiber quality.
“We’re using systems-level data and computational modeling to engineer cotton with desired cell wall characteristics,” said Szymanski, a professor in Purdue’s Department of Botany and Plant Pathology. “The first step is to clearly define the underlying regulatory circuits that control fiber cell wall properties and morphology. The next step will be to create mechanical models of the cell that, through cycles of experimenting, modeling and refinement, can predict how to engineer specified fiber traits.”
Much of this work has already been done in the model plant Arabidopsis using a similar single-cell system. Szymanski has previously mapped intracellular signaling networks onto a mechanical model of the cell to determine how cell shape is specified. Now, his team will translate those data and modeling frameworks to cotton fiber development.
The multidisciplinary team includes Purdue’s Jun Xie, a professor in the Department of Statistics; Iowa State University’s Jonathan Wendel, a distinguished professor in the Department of Ecology, Evolution, and Organismal Biology; and Olga Zabotina, a professor in the Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology.
Szymanski said the blending of biology, engineering, and statistics will be key to filtering and integrating interrelated data to make progress toward altering cotton cell shapes. Of particular importance is the finite element model used in material science for aerospace applications.
“This model is used for other materials, but it’s ideal for simulating the conditions that will be present in a thin-walled, pressurized plant cell. It’s a powerful tool, and we are driving its use in the field of plant growth control,” Szymanski said. “That modeling framework can help guide our experiments to engineer cell wall properties that control a particular trait.”
Those experiments will generate a large amount of data that statisticians like Xie will have to mine and analyze to determine what’s valuable and how to apply it to future experiments.
“This project poses so many statistical challenges. You do not know which specific proteins or genes are most important, so we have to find clever ways to leverage prior information and data integration tools to find the most important molecular players in cotton fiber development,” Xie said. “Once we determine which gene and protein interactions are important, which ones are related to the particular phenotypes we’re interested in, we can use them to make predictions and even engineer for enhanced cotton traits.”
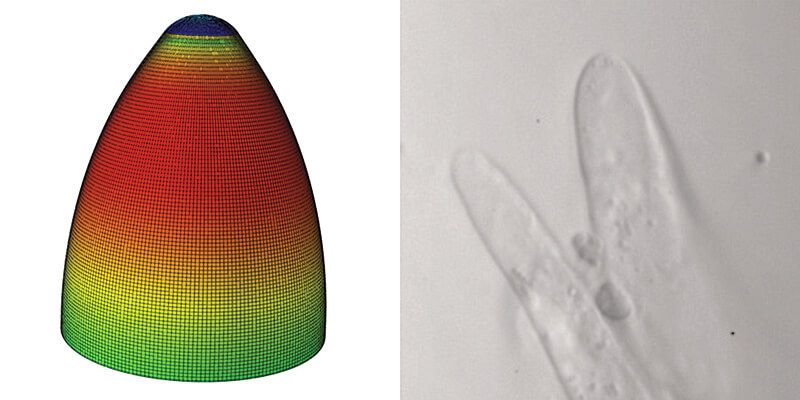 A finite element computation model of a growing cotton cell (left) simulates the shape and growth properties of the actual cotton fiber (right). Purdue’s Dan Szymanski and collaborators will work to engineer cotton fiber traits to make high-yielding U.S. varieties more similar to luxury varieties. (Photos courtesy of Dan Szymanski)
A finite element computation model of a growing cotton cell (left) simulates the shape and growth properties of the actual cotton fiber (right). Purdue’s Dan Szymanski and collaborators will work to engineer cotton fiber traits to make high-yielding U.S. varieties more similar to luxury varieties. (Photos courtesy of Dan Szymanski) 





